Cell-free DNA screening in twin pregnancies
Article information
Abstract
Cell-free DNA (cfDNA) screening for fetal aneuploidies is clinically available and exhibits better performance than conventional serum screening tests. However, data on the clinical performance of cfDNA screening in twin pregnancies are limited. In this review, we summarized the clinical performance and evaluated the feasibility of cfDNA screening in twin pregnancies based on recent studies and recommendations. The performance of cfDNA screening for trisomy 21 in twin pregnancies is similar to that in singleton pregnancies. Specifically, cfDNA screening has a higher detection rate and lower false-positive rate compared with conventional serum screening. Consequently, recent international guidelines from several academic communities have recommended that cfDNA screening for aneuploidy in twin pregnancies could be considered. Moreover, twin pregnancies can present with specific conditions, such as different zygosities and vanishing twins; therefore, individualized counseling and management are required. Further clinical studies with more twin pregnancies are required for a more accurate analysis.
Introduction
Cell-free DNA (cfDNA) screening for fetal aneuploidy (non-invasive prenatal testing) in maternal plasma has been clinically available for several years. In singleton pregnancies, cfDNA screening for common aneuploidies (trisomy 21, 18, and 13) exhibits better performance than conventional serum screening tests. A previous study reported that the detection rates (DRs) of cfDNA screening for trisomy 21, 18, and 13 were approximately 99%, 97%, and 92%, respectively, and the combined false-positive rate (FPR) was 0.04% [1]. Therefore, cfDNA screening is the most sensitive screening option for common aneuploidies [2,3].
In recent years, the incidence of twin pregnancies has increased worldwide with advancing maternal age and the increased use of assisted reproductive technology (ART) [4,5]. In 2021, multiple pregnancies were reported in association with approximately 3% and 5.4% of all births worldwide [6] and Korea [7], respectively. Twin pregnancies are generally associated with an increased incidence of complications and adverse outcomes. The risk of miscarriage due to invasive testing is higher in twin pregnancies than in singleton pregnancies [8–10]. Hence, better noninvasive screening methods with higher test performance are required in twin pregnancies to detect chromosomal abnormalities and avoid unnecessary invasive testing. Nevertheless, conventional serum screening is less accurate for twin pregnancies than for singleton pregnancies [11,12]. Combined screening has a DR of 75% and an FPR of 9% in twin pregnancies, which are lower than those in singleton pregnancies [13].
Therefore, cfDNA screening could be potentially advantageous in twin pregnancies, as it is not invasive and has a reduced risk of fetal loss [14]. Owing to the paucity of data on the clinical performance of cfDNA screening in twin pregnancies in the past, cfDNA screening was not recommended in twin pregnancies when first introduced in clinical practice. However, extensive prospective studies have reported on the performance of cfDNA screening in twin pregnancies, and the recommendations for cfDNA screening are changing. In this review, we summarize the clinical performance and evaluate the feasibility of cfDNA screening for twin pregnancies based on recent studies and recommendations.
cfDNA in twin pregnancies
An adequate level of placenta-derived or fetal cfDNA is essential for cfDNA screening. The fetal fraction (FF) was defined as the ratio of fetal to total cfDNA levels in the maternal plasma. A minimum FF of 2–4% is required for accurate cfDNA screening [15,16]. Total FF was higher in twin pregnancies than in singleton pregnancies, although the difference was less than two-fold [17–20]. Additionally, the contribution of each twin to FF is reportedly lower than that of a singleton [19,21]. For example, one study reported that fetal cfDNA levels were approximately 35% higher in twin pregnancies than in singleton pregnancies (18.1% vs. 13.4%). The study also estimated that the average effective FF for each twin pregnancy was two-thirds that for singleton pregnancies [18].
Furthermore, the FF of each twin reportedly differs based on zygosity. Struble et al. [20] reported that the median FFs of mono- and dizygotic twins were 14.0% (range, 8.2–27.0%) and 7.9% (range, 4.9–13.0%), respectively. Almost all monozygotic twins have the same genotype, and their FF is higher than that for singletons, suggesting that cfDNA screening is more feasible and effective for monozygotic twin pregnancies than for singleton pregnancies [22]. In contrast, cfDNA levels are generally lower for each dizygotic twin than for singletons, and different fetal genotypes can show different cfDNA levels [21]. Thus, describing the zygosity and FF of an individual fetus is essential for the reliable screening and interpretation of twin pregnancies.
Assessing zygosity using cfDNA screening
Studies have also evaluated different applications of cfDNA screening, such as zygosity assessment. Identifying zygosity can be helpful when chorionicity is not determined in early pregnancy [23,24]. The determination of genotypic differences is necessary for optimal cfDNA screening and interpretation.
The cfDNA levels of each dizygotic twin were analyzed separately using single-nucleotide polymorphism (SNP) analysis to determine FF and zygosity. Detecting different allele combinations between dizygotic twins is sufficient to assess zygosity, and SNP patterns can be distinguished even in fetuses with low FFs [21]. Norwitz et al. [22] used SNP analysis to distinguish zygosity with 100% sensitivity and specificity.
Performance of cfDNA screening for aneuploidy in twin pregnancies
1. Common trisomies
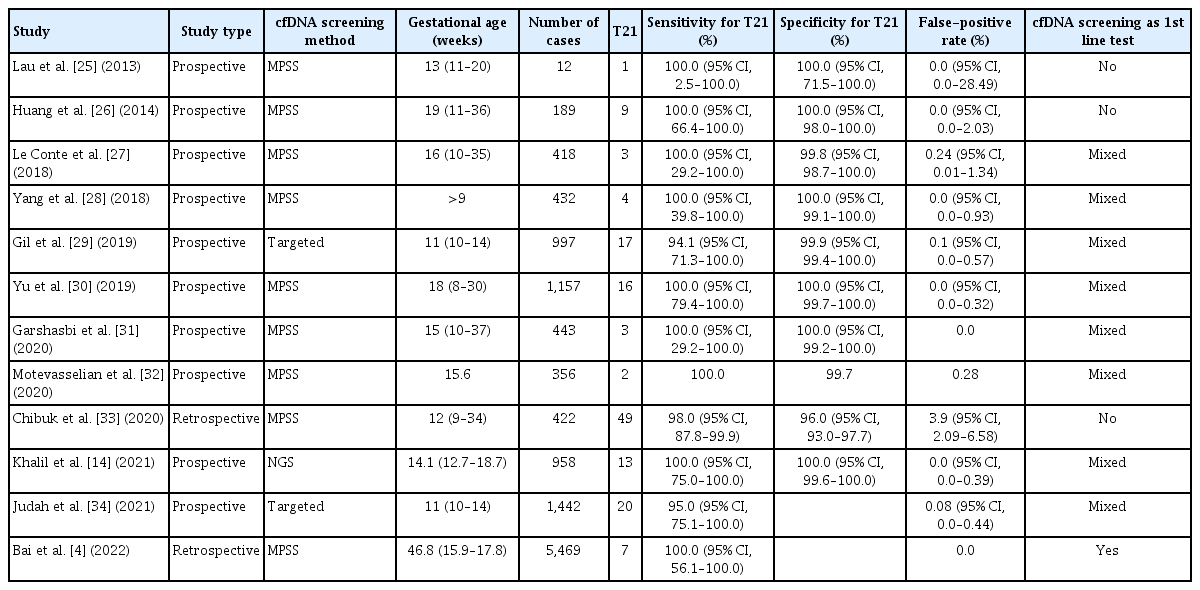
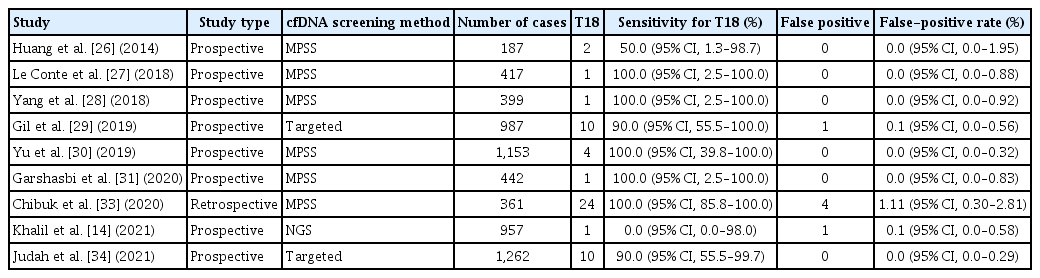
Recent studies have reported that cfDNA screening performance for trisomy 21 in twin pregnancies is similar to that in singleton pregnancies. However, cfDNA screening performance tends to be less accurate for trisomies 18 and 13. Tables 1, 2 present the clinical performance of cfDNA screening for trisomy 21 and 18 in several recent studies [4,14,25–34].
A recent meta-analysis analyzed the cfDNA screening performance in singleton and twin pregnancies. Data from twin pregnancies were insufficient compared with those from singleton pregnancies. This meta-analysis examined five prospective studies with 24 and 1,111 cases of trisomy 21 and non-trisomy 21, respectively. The DR and FPR were 100% (95% confidence interval [CI], 95.2–100%) and 0% (95% CI, 0.0–0.003%), respectively, for trisomy 21 [35]. Another meta-analysis included 10 retrospective and prospective studies with 69 cases of trisomy 21 in twin pregnancies, and the DR was 99% (95% CI, 92–100%). The meta-analysis also examined cases of trisomies 13 and 18. The DR was 85% (95% CI, 55–98%) in 13 cases of trisomy 18. In addition, three cases of trisomy 13 and 2,008 twin pregnancies with nontrisomy 13 were studied; the DR and FPR were 100% (95% CI, 30–100%) and 0.05%, respectively [36].
Gil et al. [29] conducted a meta-analysis using one self-study and seven other studies with a total of 56 trisomy 21 and 3,718 non-trisomy 21 twin cases. The pooled weighted DR was 98.2% (95% CI, 83.2–99.8%), and the FPR was 0.05% (95% CI, 0.01–0.26%) for trisomy 21 cases. The DR and FPR varied between 94.1% and 100% and between 0% and 0.24%, respectively, in the included studies. A total of 18 cases of trisomy 18 and 3,143 cases of non-trisomy 18 were combined in five studies, and the pooled weighted DR and FPR were 88.9% (95% CI, 64.8–97.2%) and 0.03% (95% CI, 0–0.33%), respectively. The DR in individual studies varied between 50% and 100%, and the FPR varied between 0% and 0.10% for trisomy 18 cases. Only three cases of trisomy 13 were included in this meta-analysis. Furthermore, three trisomy 13 cases and 2,569 non-trisomy 13 cases were included; the DR and FPR were 66.7% (2/3) and 0.19% (5/2,569), respectively [29]. Notably, this study suggests that the performance of cfDNA screening for trisomy 21 may be similar between twin and singleton pregnancies. The same study group conducted a meta-analysis of 1,963 cases of trisomy 21 and 223,932 singleton pregnancies with non-trisomy 21. The pooled weighted DR and FPR were 99.7% (95% CI, 99.1–99.9%) and 0.04% (95% CI, 0.02–0.07%), respectively [35].
Khalil et al. [14] conducted a meta-analysis of 11 large-scale prospective multicenter studies and one self-study. The DR was 95% (95% CI, 90–99%) in 74 cases of trisomy 21, and the FPR was 0.09% (95% CI, 0.03–0.19%) in 5,598 euploid pregnancies. The DR and FPR ranged from 94.1% to 100% and from 0% to 0.24%, respectively, in individual studies. A total of 22 cases of trisomy 18 and 4,869 twin pregnancies with non-trisomy 18 were recorded, and the DR and FPR rates were 82% (95% CI, 66–93%) and 0.08% (95% CI, 0.02–0.18%), respectively. In addition, the DR and FPR ranged from 50% to 100% and from 0% to 0.10%, respectively, in each study. The DR and FPR were 80% and 0.13% in 5 trisomy 13 cases and 3,881 euploid cases, respectively [14].
cfDNA screening in twin pregnancies showed a high performance rate and DR and low FPR, which were comparable to those in singleton pregnancies, especially for trisomy 21 screening. The higher screening performance of cfDNA in twin pregnancies is advantageous as it reduces miscarriage risk and the need for unnecessary invasive testing [21,29]. Some studies have reported a lower invasive testing rate after cfDNA screening [4,37]. However, the number of cases of trisomies 18 and 13 was relatively small for a precise evaluation of the clinical performance of cfDNA screening. Therefore, further studies are warranted.
Previously, cfDNA screening in twin pregnancies was not strongly recommended because of a lack of data. Thus, before 2018, many academic societies did not recommend cfDNA screening in twin pregnancies but rather requested further investigation [38]. However, various studies have reported that the clinical performance of cfDNA screening in twin pregnancies is superior to that of conventional serum screening tests, and the accuracy of cfDNA screening is comparable between singleton and twin pregnancies [17,27,29,36,39]. Based on these results, recent international academic community guidelines have recommended cfDNA screening for aneuploidy during twin pregnancies. For example, the American College of Obstetricians and Gynecologists (ACOG), International Society for Prenatal Diagnosis (ISPD), American College of Medical Genetics and Genomics (ACMG), and Royal College of Obstetricians and Gynecologists now support cfDNA screening in twin pregnancies [40–43]. These communities recommend cfDNA screening for trisomy 21. As a level B recommendation, the ACOG approved cfDNA screening for twin pregnancies [40]. The ISPD also recommends moderate application of cfDNA screening for common trisomies in twin pregnancies based on high DR, low FPR, and high predictive values [41]. Additionally, ISPD recommends that laboratories consider zygosity when interpreting all test results and FFs [41]. The ACMG recently recommended cfDNA screening over traditional screening methods for common trisomies in twin pregnancies [42]. However, several researchers have encouraged further studies to verify the performance of cfDNA screening.
2. Sex chromosome abnormalities
Data on the performance of cfDNA screening for sex chromosome abnormalities (SCA) are limited. However, some studies have demonstrated the potential of cfDNA screening for common SCA in twin pregnancies. Bai et al. [4] identified four cases of SCA with 100% sensitivity (95% CI, 39.6–100%) and 99.8% specificity (95% CI, 99.5–99.9%) for cfDNA screening. The positive predictive value (PPV) was 40% (95% CI, 13.7–72.6%), and the negative predictive value was 100% (95% CI, 99.8–100%), which were similar to those in singleton pregnancies [4]. Nevertheless, the performance of cfDNA screening for SCA is limited to twins, depending on the technology used for analysis and chorionicity. Thus, further large-scale studies are required to evaluate the performance of cfDNA screening [42].
3. Test failure
Although cfDNA screening is clinically feasible in twin pregnancies, the test failure rate is high in twin pregnancies [44]. The initial cfDNA screening failure rate in twin pregnancies ranged from 1.6% to 13.2% (median, 3.6%) [38]. Another study reported initial test failure rates of 3.4%, 4.9%, and 11.3% in singleton, monochorionic twin, and dichorionic twin pregnancies, respectively [45].
Low FF in twin pregnancies increases the test failure rate [46] and false-negative rates with cfDNA screening [28]. In addition, fetuses with trisomy are likely to have fewer FFs. If only a dizygotic twin fetus had aneuploidy, a normal co-twin would have a higher contribution to the total FF, which can produce false-negative results [14]. Therefore, the minimum FF requirement may be higher in twin pregnancies [14].
Furthermore, other contributing factors have been reported to increase the test failure rates. Bai et al. [4] reported a test failure rate of 1.2% (32/2,671) due to a low FF. Test failure in this study tended to be more frequent among women with a history of ART and a higher body mass index (BMI). The FF was significantly lower in women with ART history (odds ratio [OR], 2.89; 95% CI, 1.32–6.29%) and higher BMI (OR, 1.17; 95% CI, 1.07–1.29%) [4]. Another study reported that increased maternal weight, ART history, dichorionicity, primiparity, and low gestational age were essential contributors to cfDNA screening test failure [45]. Adipocyte turnover increases with increasing BMI and maternal cfDNA levels, resulting in decreased FF [47]. The FF was found to decrease by 0.541% for every 1 kg/m2 increase in BMI [48]. Impaired placentation is a possible cause of cfDNA screening failure in pregnant women requiring ART [4].
Further genetic counseling, comprehensive ultrasound evaluation, and diagnostic testing are recommended when test failure is reported after cfDNA screening [38,49]. However, ISPD suggests that a second blood sample draw can be considered if there is sufficient time [41].
Vanishing twin (VT) pregnancies
After ART, 10–39% of twin pregnancies demonstrate VT [50]. VT is associated with adverse perinatal outcomes and increased fetal aneuploidy rate [51,52]. The cfDNA of VTs can remain in the maternal plasma for 8–15 weeks and can be detected via cfDNA screening [52,53]. Chromosomal abnormalities, including trisomies, are the primary causes of miscarriage and VT. Although VT affects maternal serum markers, the status of serum markers in pregnant women with VT remains uncertain [54]. However, the remaining cfDNA of affected and demised fetuses can influence the cfDNA screening results, thereby increasing the FPR [52]. One study examined 847 pregnant women with VT who underwent cfDNA screening for common trisomies. The PPVs for trisomies 21, 18, and 13 were 50% (6/12), 11.1% (1/9), and 0% (0/6), respectively. PPV was lower in pregnancies with VT than in those without VT [50]. Moreover, Balaguer et al. [53] reported a higher screening positivity rate (5.8% vs. 2.5%; P<0.01) and FPR (2.6% vs. 0.3%; P<0.01) in pregnancies with VT than in singleton pregnancies. However, the study also reported that the positive screening rate (3.1%) and FPR (0.8%) decreased when cfDNA screening was performed after 14 weeks of gestation. Hence, the authors suggested that cfDNA screening could also be performed in pregnant women with VT after 14 weeks of gestation [53]. However, further studies are required to confirm these findings. Therefore, the ACOG recommends informing patients with VT of the possible inaccuracy of cfDNA screening results and offering diagnostic testing [40].
Conclusion
Recent studies, including several meta-analyses, have reported good cfDNA screening performance for common aneuploidies in twin pregnancies. Owing to its lower invasiveness, higher DR, and lower FPR, cfDNA screening has superior performance compared to conventional serum screening, and its performance of cfDNA screening is comparable between twin and singleton pregnancies [17,27,36,39,55]. A critical advantage of cfDNA screening is its low FPR, which reduces the need for invasive diagnostic tests. The results of extensive prospective studies analyzing the clinical performance of cfDNA screening have been reported, and recommendations regarding the application of cfDNA screening are changing. Recent recommendations and guidelines of the ACOG, ISPD, and ACMG support the use of cfDNA screening as the primary screening method for common fetal aneuploidies in twin pregnancies. Therefore, physicians should explain cfDNA screening for common aneuploidies during counseling of women with twin pregnancies. Conventional serum or cfDNA screening should be implemented as necessary or according to maternal preferences. However, more data are required to ascertain the utility of cfDNA screening for SCA owing to its low prevalence. Recent studies have demonstrated the effectiveness of cfDNA screening for SCA during twin pregnancies [4].
Twin pregnancies are more predisposed to complications (such as miscarriages and adverse pregnancy outcomes) than singleton pregnancies. Moreover, women with twin pregnancies can present with specific conditions such as different zygosities and VT. Therefore, cfDNA screening may be more complicated in twin pregnancies. Physicians should analyze individualized situations and provide appropriate counseling. Further clinical studies including a larger sample of women with twin pregnancies are required for a more accurate analysis.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Ethical approval
This study did not require approval from the Institutional Review Board because no patient data were included. This study was conducted in accordance with the principles of the Declaration of Helsinki.
Patient consent
Written informed consent and the use of patient images were not required for publication.
Funding information
None.