Optimization to detect TP53 mutations in circulating cell-free tumor DNA from patients with serous epithelial ovarian cancer
Article information
Abstract
Objective
Circulating cell-free tumor DNA (cfDNA) is the DNA released by apoptotic and necrotic cells of the primary tumor into the blood during the period of tumor development. The cfDNA reflects the genetic and epigenetic alterations of the original tumor. TP53 mutations are a defining feature of high-grade serous ovarian carcinoma. We optimized the methods for detecting TP53 mutations in cfDNA from blood samples. We confirmed the correlation of TP53 mutation in primary ovarian cancer tissue and it in cfDNA using digital polymerase chain reaction (dPCR).
Methods
We found 12 frequent mutation sites in TP53 using The Cancer Genome Atlas and Catalogue of Somatic Mutations in Cancer data and manufactured 12 primers. The mutations in tissues were evaluated in fresh-frozen tissue (FFT) and formalin-fixed paraffin-embedded tissue (FFPET). We performed a prospective analysis of serial plasma samples collected from 4 patients before debulking surgery. We extracted cfDNA and calculated its concentration in blood. dPCR was used to analyze TP53 mutations in cfDNA, and we compared TP53 mutations in ovarian cancer tissue with those in cfDNA.
Results
Ten primers out of 12 detected the presence of TP53 mutations in FFT, FFPET, and cfDNA. In FFT and FFPET tissue, there were no significant differences. The average cfDNA concentration was 2.12±0.59 ng/mL. We also confirmed that mutations of cfDNA and those of FFT were all in R282W site.
Conclusion
This study developed detection methods for TP53 mutations in cfDNA in ovarian cancer patients using dPCR. The results demonstrated that there are the same TP53 mutations in both ovarian cancer tissue and cfDNA.
Introduction
Ovarian cancer comprises the majority of malignant ovarian tumors in women aged 50 to 70 years. However, because there is no specific biomarker for the diagnosis, prognosis, or response to treatment of ovarian cancer, most cases are diagnosed in advanced stages. Better diagnosis strategies for early detection are thus needed to improve survival rates of women with ovarian cancer.
The Cancer Genome Atlas (TCGA) study has reported that TP53 mutations are the most common (96%, 302 sites) in epithelial ovarian cancer. Frequently mutated “hot spots” have been found at the R175, G245, R428, R273, and R282 domains [12]. TP53 mutations are thought to reflect the disease status of ovarian cancer patients, such as stage, risk of relapse, and response to treatment. However, due to the invasive nature of ovarian cancer tissue collection, studies on TP53 mutations in ovarian cancer are limited. To address this hurdle, researchers have begun studying circulating cell-free tumor DNA (cfDNA).
The cfDNA was discovered in the cell-free fraction of blood by Mendel in 1948. Initially, cfDNA attracted little attention from scientists; however, in 1994 mutated RAS gene fragments were found in the blood of cancer patients, shedding light on the importance of cfDNA [345]. Because cfDNA consists of tumor-specific sequence alterations, it is expected to reflect diagnosis, response to treatment, and prognosis of cancer patients. Mutations have been in cfDNA from various cancers, such as pancreatic, colon, breast, and lung cancer. Most cfDNA consists of double-strand DNA and nucleus-protein complex [6]. Because cfDNA is released in blood from various malignant origins, mutations found in cfDNA reflect the genetic characteristics of the primary or metastatic tumor. Recently, many studies are focused on quantifying cfDNA in order to characterize primary cancers. However, only a very small amount of cfDNA is found in blood, making it difficult to detect cfDNA using quantitative real-time polymerase chain reaction (PCR). Digital PCR (dPCR) has many potential advantages over real-time PCR, including the ability to obtain absolute quantification without external references. This technique has higher sensitivity, precision in PCR efficiency, and high day-to-day reproducibility than conventional PCR [7].
The primary aim of this study was to optimize the method for detecting cfDNA mutations in blood acquired from epithelial ovarian cancer patients. A secondary aim was to confirm if TP53 mutations in tissues and cfDNA were correlated with each other.
Materials and methods
1. Analysis of TP53 mutations and selection of high frequency positions
We analyzed high frequency TP53 mutations sites using TCGA, the Catalogue of Somatic Mutations in Cancer (COSMIC) data (http://cancergenome.nih.gov/, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) [89].
2. Primer design and confirmation of TP53 mutations in cancer tissue
We referred to Life Technologies Inc. (Carlsbad, CA, USA) to manufacture Taqman® probes to search the 12 selected sites (Table 1). Twelve primer sets were designed for detection of TP53 mutations; however, only 11 primer sets were made, while a primer set for V157F was not available. We performed real-time PCR in fresh-frozen tissue (FFT) and formalin-fixed paraffin-embedded tissue (FFPET) using manufactured probes. PCR reactions were carried out in an Applied Biosystem® GeneAmp® PCR System 9700 (Life Technologies Inc.). After denaturation at 95°C for 15 seconds, annealing was performed at 60°C for 60 seconds (45 cycles, amplification).
3. Measurement of cfDNA concentration in whole blood by double-stranded DNA (dsDNA) assay and dPCR
There are no standard methods for detecting cfDNA in ovarian cancer. We therefore aimed to prepare blood sampling and detect cfDNA using dPCR. After sampling blood, we separated plasma from whole blood. We measured the average concentration of cfDNA using Qubit™ dsDNA assay kits (Invitrogen, Carlsbad, CA, USA). We then performed PCR using the Applied Biosystems® GeneAmp® PCR System 9700 (Life Technologies Inc.). The conditions were as follows: denaturation at 94°C for 30 seconds, annealing at 60°C for 1 minute, followed by 40 cycles of amplification.
4. Detection of TP53 mutations in tissues and blood from ovarian cancer patients
Patient inclusion criteria were as follows: female patients over the age of 20, patients with ovarian cancer or suspected ovarian cancer, those who were scheduled surgery for a pelvic mass, and those with post-surgery pathologic findings indicating papillary serous ovarian cancer. Patient with diagnosed malignant cancer over the past 10 years (except for non-melanoma skin cancer); those with clinically significant infectious disease or immune-compromised state; those who underwent surgical treatment, open surgical biopsy, or suffered significant trauma within 28 days; those with serious wounds, ulcers, and fractures; those with significant medical problems, such as uncontrolled cardiovascular, nervous system, and endocrine disorders or mental illness; those with metastatic ovarian cancer; and those who received blood transfusion of more than 450 mL within 28 days of screening were excluded from the study
5. Blood and tissue sampling
Patients who visited Asan Medical Center were included in this clinical study. The study was approved by the Internal Review Board of Asan Medical Center (approval number: 2011-0688) and all patients provided informed consent prior to enrollment into the study.
1) Tissue collections and DNA extraction
During surgery, we collected 1 g of ovarian cancer tissue and placed it immediately in storage tubes at −80ºC until further use. After 10 mg of frozen tissue was chopped, 200 µL 4 M TNES-Urea and 10 µL EDT were added and mixed in a 55ºC water bath until fully dissolved. Then, 180 µL LDT was added over 15 seconds and incubated at 70ºC for 10 minutes. The mixture was centrifuged, 250 µL 99% ethanol was added, and the mixture was again centrifuged. DNA was extracted using QuickGene® (Kurabo Industries, Osaka, Japan).
2) Blood sampling
Samples of blood were obtained from the patient's vein before debulking surgery. After blood sampling, the tubes were inverted.
3) Plasma separation
Plasma preparation was conducted at room temperature and completed within 2 hours after blood collection. In order to separate blood components, the tube was allowed to stand upright for 60 minutes. Blood samples with clear layer separation were centrifuged for 15 minutes. The supernatant was transferred to a sterile tube and then centrifuged at 1,600 g for 10 minutes at room temperature. The supernatant was transferred to a fresh tube until further testing.
4) cfDNA extraction
We extracted cfDNA using the QIAmp Circulating Nucleic Acid kit® (Qiagen, Hilden, Germany). Qiagen proteinase K® (Qiagen; 500 µL) was placed in a 50-mL tube, followed by the addition of 4 mL ACL buffer and 5 mL plasma; the mixture was mixed for 30 seconds and then maintained at 60°C for 30 minutes, followed by the addition of 3.6 mL of ACB buffer. The mixture was kept on ice for 5 minutes; then 600 µL ACW1 buffer 600 was added using a vacuum pump of a QIAvac 24 Plus® (Qiagen). ACW2 (750 µL) buffer and 100% ethanol (750 µL) were sequentially passed through the column. The tube was centrifuged for 3 minutes, and then left for 10 minutes at 56°C with an open lid to dry completely. AVE buffer (50 µL) was carefully added to the middle of the film, allowed to stand at room temperature for 3 minutes, and then centrifuged for 1 minute at 14,000 rpm to obtain the DNA.
5) Detection TP53 mutation using dPCR
We detected cfDNA mutations in TP53 using the QX100TM Droplet Digital™ PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). The reactions were optimized with respect to the primer between 250 nM and 900 nM). No template controls (NTCs) were as negative controls, which monitored contaminations and primer-dimer formation [10]. Briefly, 10 ng cfDNA, probe, and droplet digital PCR (ddPCR) reagent (Bio-Rad Laboratories Inc.) were mixed. The DNA mixture and oil were placed into the droplet generation cartridge. We performed PCR using the Applied Biosystems® GeneAmp® PCR System 9700 (Life Technologies Inc.) under the following conditions: denaturation at 94°C for 30 seconds and annealing at 60°C for 1 minute followed by 40 cycles of amplification. TP53 mutations were identified using ddPCR reader (Bio-Rad Laboratories Inc.).
Results
1. Mutational sites for TP53 using data from the TCGA, COSMIC experiment
TP53 mutations associated with ovarian cancer have been found in 302 sites, 187 (61.9%) of which are missense mutations [8]. We selected the 12 most frequent sites for TP53 mutations, which are the sites for 23.18% of all mutations (Table 1).
2. Patient characteristics
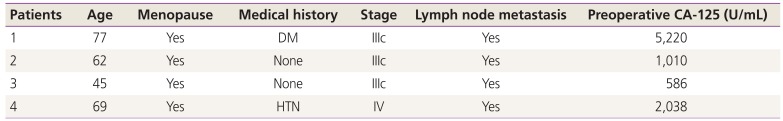
We collected ovarian cancer tissues from 4 patients to determine TP53 mutations in tissues. The mean age of the patients was 63.25 years, and all patients were postmenopausal women. Three patients had stage IIIc ovarian cancer, and one patient had stage IV disease. Preoperative cancer antigen (CA) 125 levels ranged from 172 to 5,220 U/mL (Table 2).
3. TP53 mutations in FFT and FFPET
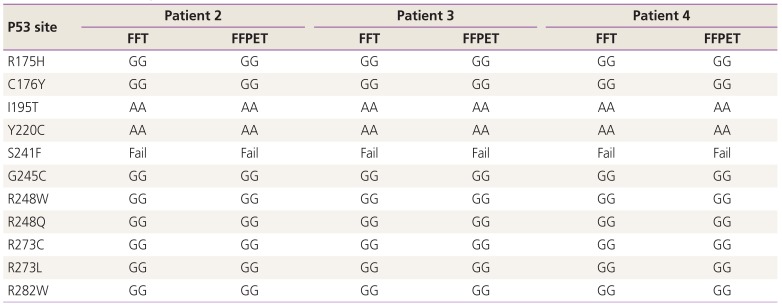
We identified TP53 mutations using FFT and FFPET in 3 patients. FFPET was not prepared for one patient. In addition, one set of primers (S241F) did not recognize the target sequence. In the results, we confirmed that 10 primers worked well and detected 61 sites (20.2%) of TP53 mutation. And, we found that there was a mutation in R282W site with one patient. The same results were found in both FFT and FFPET (Table 3).
4. Blood cfDNA concentration and detectable quantity using dPCR
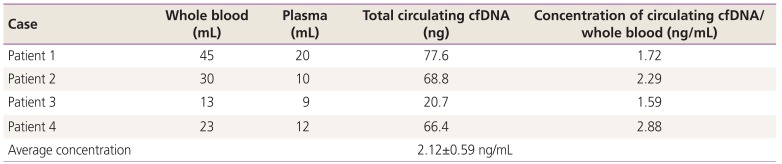
We calculated the average concentration of cfDNA obtained in order to determine the minimal amount of blood needed from the ovarian cancer patients. The amount of whole blood ranged from 13 to 45 mL, from which the amount of plasma extracted ranged from 9 to 20 mL. We confirmed that average concentration of plasma was 0.5 mL per 1 mL of whole blood. The average concentration of extracted cfDNA was 2.12±0.59 ng per 1 mL of whole blood. The average concentration of extracted cfDNA ranged from 1.59 to 2.88 ng/mL (Table 4). We performed dPCR according to the amount of cfDNA to verify the amount of detectable cfDNA. Less of 1 ng cfDNA showed false-negative results in dPCR. More than 5 ng of cfDNA produced consistent results (Table 5). Based on the results of this experiment, we set 20 mL of whole blood and 10 ng of cfDNA as the appropriate amounts for detection using dPCR.
5. Relevance of TP53 mutation in tissue and cfDNA
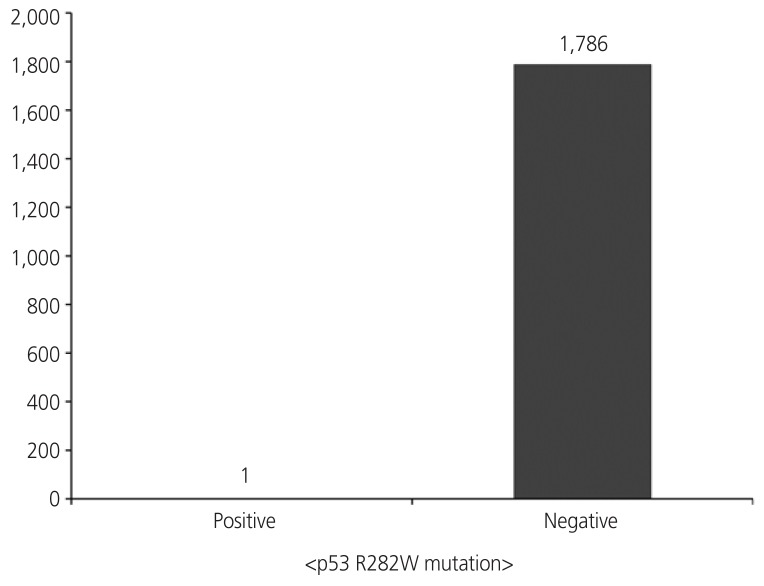
We investigated the TP53 R282W mutation in cfDNA in ovarian cancer tissue. Lime green visualization by dPCR indicated the presence of a R282W mutation. The values for the positive and negative results were 1 and 1,786, respectively, by dPCR (Fig. 1).
Discussion
Ovarian cancer is a gynecologic cancer with a high mortality rate. Currently, platinum and paclitaxel remain the only treatments, with no improvements in therapy seen over the last 20 years. In order to determine the etiology of ovarian cancer, TCGA and COSMIC studies were conducted to analyze the genetic characteristics of ovarian cancer. According to these studies, TP53 mutations were found to be responsible for 96% of somatic mutations in ovarian cancer [8]. Further studies focusing on TP53 mutations and ovarian cancer are underway. In addition, since the discovery that the TP53 gene is involved in drug treatment response, a number of studies have been conducted on TP53-related cancer treatment [11]. Specifically for ovarian cancer, a study on the involvement of TP53 mutations on the resistance to platinum treatment and cancer metastasis has been reported [12].
Studies on the relationship between TP53 mutations and treatment response are limited because of the invasive nature of tissue collection. Although it is possible to detect various TP53 mutations at once from tissue biopsy samples, it is more invasive than blood sampling and difficult to perform in a repeated manner. Also, the goal of identifying mutations in tissues is not to predict recurrence in advance or to expect the effects of treatment early — rather, biopsy results are more retrospective in nature. Cost for getting cancer tissue and testing mutations is more expensive than drawing blood as well. To address this issue, cfDNA has been used in cancer research. Because cfDNA can be measured in blood, it is much less invasive than surgery or biopsy and is faster than other procedures. Checking for residual cancer, monitoring treatment, and following up for detection of recurrence after treatment are feasible because tests for genetic and epigenetic mutations can be repeated during the treatment period. Development of new testing methods for ovarian cancer that are less invasive than surgery and easy to repeat are needed due to the absence of a hematological and radiological method to diagnose ovarian cancer and to accurately determine disease progress.
Current detection methods to verify cfDNA mutations are limited because the cfDNA quantity found in blood is insufficient and because cfDNA has unstable characteristics compared to tissue DNA. Diaz and Bardelli [13] described that in advanced tumors, digital genomic approaches have high sensitivity, with the mutation identified in the tumor tissue matching the mutation in the cfDNA fraction in virtually every case. These techniques have expanded the ability for detecting a single point mutation, and now multiple genes of interest can be investigated in one sample. Amplifications, rearrangements, and aneuploidy can now be detectable as well [13]. Therefore, cfDNA can be detected by dPCR, which has gained attention due to its high accuracy and reproducibility. Compared to conventional PCR, dPCR exhibits increased sensitivity because the reaction is processed by diluting it in a very small space to digitize, while offering high reproducibility as well [10]. However, this technology is not yet applicable to various fields.
To the best of our knowledge, our present report is the first prospective study on the evaluation of cfDNA mutations in patients with ovarian cancer using dPCR. Mutations were verified using various amounts of cfDNA due to the absence of a standard protocol to detect cfDNA mutations using dPCR. Because mutations can be detected from 5 ng or more of cfDNA, 10 ng of cfDNA per mutation was used to detect TP53 mutations in this study. In addition, 2.12±0.59 ng of cfDNA per 1 mL of whole blood on average was confirmed as an adequate amount for further tests. These steps allowed us to determine the concentration of cfDNA in whole blood and to identify specific mutations. Through this process, dPCR was verified as a method to screen cfDNA and can be applied in future studies.
The cfDNA is rarely applied in clinical practice as of now. However, substantial progress has been made in the discovery of new biomarkers, such as activating mutations in the epidermal growth factor receptor and the kirsten rat sarcoma viral oncogene homolog (KRAS) gene, as well as amplification of the v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2) and the echinoderm microtubule-associated protein like 4-anaplastic lymphoma receptor tyrosine kinase (EML4-ALK) fusion gene. Cancers harboring these mutations are responsive to specific targeted inhibitors [14]. The cfDNA has been reported to be a marker for the early diagnosis of pancreatic and lung cancer in the absence of another method for early diagnosis [1516]. For example, Bordi et al. [17] investigated whether anaplastic lymphoma kinase (ALK) and KRAS mutations are associated with acquired resistance to crizotinib in ALK-positive non-small cell lung cancer (NSCLC). They detected ALK and KRAS mutations in cfDNA using dPCR, and concluded that ALK acquired mutations can be detected in plasma and could represent a promising tumor marker for response monitoring [17]. Also, there is a study on lorlatinib versus crizotinib in first line treatment of patients with ALK-positive NSCLC in “clinicaltrials.gov” [18]. In this study, lorlatinib and crizotinib are used as target therapy drugs for NSCLC, tumor tissue biomarkers and peripheral blood cfDNA biomarkers are used as biomarkers.
The cfDNA can be used for the diagnosis of ovarian cancer based on the finding that its levels in the blood of an ovarian cancer patient is higher than that in healthy people. Wimberger and colleagues [19] showed a correlation between the amount of cfDNA measured before surgery and prognosis after surgery in ovarian cancer patients. These authors also reported that the total amount of cfDNA was lower after anticancer treatment. Previous studies showed that the total amount of cfDNA in ovarian cancer patients is associated with early detection and treatment response. Shao et al. [20] described that there was a correlation between the levels of serum cfDNA and the occurrence and development of ovarian cancer in patients.
However, Zhou et al. [21] suggested cfDNA as the diagnostic marker for ovarian cancer via systematic review and meta-analysis. According to their review, current evidence suggests that quantitative analysis of cfDNA has unsatisfactory sensitivity but acceptable specificity for the diagnosis of ovarian cancer, and that further large-scale prospective studies are required to validate the potential applicability of using circulating cfDNA alone or in combination with conventional markers as diagnostic biomarker for ovarian cancer, and to explore potential factors that may influence the accuracy of ovarian cancer diagnosis [2021]. In our present study, 2.12±0.59 ng of cfDNA per 1 mL of whole blood was detected. We detected various cfDNA levels (range of 1.59–2.88 ng per 1 mL). The total amount of cfDNA can increase with increasing metabolism because of infection or other causes, making it difficult to use the amount of cfDNA to determine diagnosis. The relationship between the total amount of cfDNA and the early diagnosis of ovarian cancer has not yet been clarified. Furthermore, the relationship between cfDNA and CA-125 level or disease stage remains unclear and warrants further investigation.
In our current study, an ovarian cancer-specific test was conducted to confirm TP53 mutations in cfDNA rather than comparing the total amount of cfDNA. Mutations in cfDNA are widely found in various cancers, such as colorectal, pancreatic, breast, and lung cancer [1122]. According to a study conducted by Dawson and colleagues in 2013 [23], 29 of 30 patients (97%) with a mutation in PIK3CA or TP53 in breast cancer cells showed a similar mutation in cfDNA. In fact, cfDNA measurement showed higher sensitivity than CA 15-3 and circulating tumor cells in identifying mutations in patients with metastasized breast cancer, and cfDNA closely reflected tumor size and treatment response. In our present study, 61 mutations (20.2%) were confirmed from a total of 302 TP53 mutations that appeared in TCGA. Furthermore, 1 of 4 patients we analyzed had a TP53 mutation present in the tissue, and TP53 mutations at the same location were observed from the cfDNA of patients who showed mutations in tissue samples. Further studies are needed to elucidate the relationship between ovarian cancer tissue and cfDNA mutations. In addition, the amount of cfDNA after surgery, during anticancer treatment, and after anticancer treatment should be further investigated, as well as changes in patient mutational status through follow-up studies.
Parkinson et al. [24] retrospectively analyzed TP53 mutations in cfDNA as biomarker of treatment response for patients with response for patients with relapse high-grade serous ovarian carcinoma. TP53 mutant allele fraction correlated with volumetric measurements (Pearson r=0.59; P<0.001), and this correlation increased when patients with ascites were excluded (r=0.82). In nearly all relapsed patients with disease volume >32 cm3, cfDNA was detected at ≥20 amplifiable copies per milliliter of plasma. Response to chemotherapy with cfDNA was seen earlier than with CA-125. The authors demonstrated that cfDNA is correlated with volume of disease at the start of treatment in women with high-grade serous ovarian carcinoma and that a decrease of ≤60% in TP53 mutant allele fraction after one cycle of chemotherapy was associated with shorter time to progression. These results provide evidence that cfDNA has the potential to be a highly specific early molecular response marker in high-grade serous ovarian carcinoma [24]. However, this study limited in its retrospective design, small sample size, and heterogeneity of treatment within the cohort. We are thus planning a prospective study that uses a substantially larger sample size for assessing the utility of cfDNA as biomarker.
The main limitation of our study is its small sample size. Because the main objective of this study was to optimize experiments to extract cfDNA from blood and to identify its mutations prior to application to many patients, we prospectively selected 4 cases to optimize the experimental method. These 4 cases were enough to establish the detecting method for cfDNA. Also, our current study showed the potential clinical value of cfDNA. Based on our current findings, we expect that following research will benefit from investigating whether cfDNA level is related with progression and treatment of disease. We also aim to test whether cfDNA has meaningful clinical value as a personalized biomarker.
Acknowledgements
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI06C0868).
Notes
Conflict of interest: No potential conflict of interest relevant to this article was reported.